Currently, I am a Postdoctoral Scholar at the State Key Laboratory of Membrane Biology, Beijing Tsinghua Institute for Frontier Interdisciplinary Innovation, collaborating closely with Prof.Qiangfeng Cliff Zhang.
My research centers on cryo-electron microscopy (cryo-EM) structure determination and discovery. I am broadly interested in data-driven algorithm design and deep learning applications across multiple scales—from real-world 2D/3D computer vision and graphics to biological micro-world challenges such as cryo-EM density map-based molecular structure determination.
Prior to this, I served as a Postdoctoral Scholar at Tsinghua University, where I collaborated closely with Professor Qiangfeng Cliff Zhang. During my Ph.D. study period, my work focused on computational methods for structural biology and biomolecular data analysis.
Earlier, as a visiting scholar at the Chinese University of Hong Kong, I worked with Professor Xiaogang Wang and closely collaborated with Zhe Wang, Jianping Shi, and Hongsheng Li on projects related to 3D object detection and molecular structure modeling..
I received my Ph.D. degree in School of Life Sciences of Tsinghua University in 2021. Before I came to Tsinghua University, I received a master and bachelor degree in Computer Science in Tianjin Polytechnic University in 2016 and 2013, respectively.
Feel free to say hi: xukui.2016 at tsinghua.org.cn
What's New
[Jan 2026] Two paper accepted to ICLR, congratulations to Weining, Kai, Fuyao, and Xiaozhu.
[Jun 2025] CryoNet.DB was published in NCAIB.
[Mar 2025] One paper accepted to AAAI, congratulations to Muzhi.
[Dec 2024] One paper accepted to PNAS, congratulations to Tongtong and Wenze.
[Oct 2024] One paper accepted to PNAS, congratulations to Tongtong and Zhangqiang.
[May 2023] One paper accepted to Nucleic Acids Research, congratulations to Yiran and Lei.
[Apr 2023] One paper accepted to Cell, congratulations to Prof Gao and Li.
[Mar 2023] One paper accepted to Journal of Molecular Biology, congratulations to Muzhi.
[Nov 2022] Our PrismNet method paper won the 2021 Sanofi-Cell Research Outstanding Research Article Award.
[Feb 2022] Our PrismNet method paper was awarded as the Top Ten Advances of 2021 in Bioinformatics in China by Genomics, Proteomics and Bioinformatics (GPB).
[Oct 2021] One paper accepted to Nature Machine Intelligence, congratulations to Dr. Jing Gong.
[Jul 2021] CryoNet (https://cryonet.ai) is online!
[Jan 2021] One paper accepted to Science. Proud to apply our improved A2-Net to identify new proteins in Spliceosome,
congratulations to Dr. Rui Bai and Dr. Ruixue Wan.
[Jan 2021] Our PrismNet method paper accepted to Cell Research, congratulations to all the team members.
[Nov 2020] Our PrismNet Prediction on SARS-CoV-2 paper accepted to Cell, congratulations to Dr. Lei Sun, Pan Li and Wenze.
[Oct 2020] One paper accepted to Nucleic Acids Research, congratulations to Pan Li and Xiaolin.
[Jul 2020] One paper accepted to Bioinformatics.
[Jul 2020] Proud to apply our PrismNet to predict host cell target proteins bind to SARS-CoV-2 RNA! Paper out on BioRxiv.
[May 2020] PrismNet method paper out on BioRxiv: Predicting dynamic cellular protein-RNA interactions using deep learning and in vivo RNA structure,
code are available at Github.
[Jan 2020] One paper accepted to ICRA 2020, congratulations to Hongwei Yi.
[Oct 2019] Paper out on Nature Communications: SCALE method for single-cell ATAC-seq analysis via latent feature extraction, code are available at SCALE.
[Mar 2019] Paper out on BioRxiv: VRmol: an Integrative Cloud-Based Virtual Reality System to Explore Macromolecular Structure, code are available
at VRmol.
[Jan 2019] Presented our work A2-Net at AAAI 2019, Hawaii.
Research
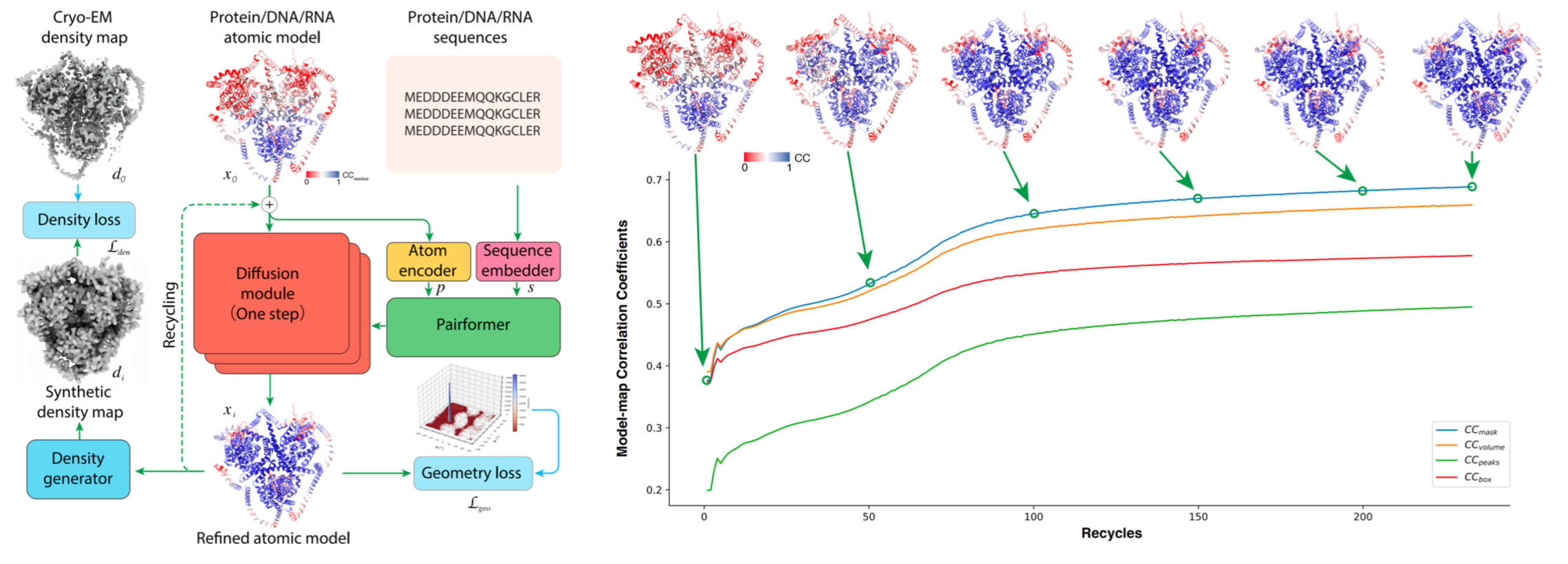
CryoNet.Refine: A One-step Diffusion Model for Rapid Refinement of Structural Models with Cryo-EM Density Map Restraints
Fuyao Huang*, Xiaozhu Yu*, Kui Xu#, and Qiangfeng Cliff Zhang#
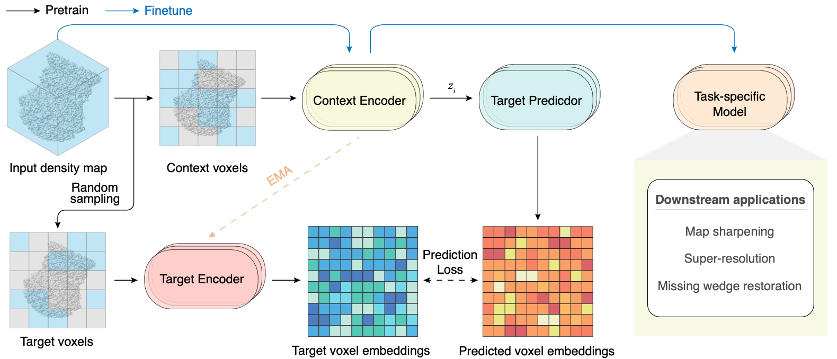
CryoLVM: Self-supervised Learning from Cryo-EM Density Maps with Large Vision Models
Weining Fu*, Kai Shu*, Kui Xu#, and Qiangfeng Cliff Zhang#
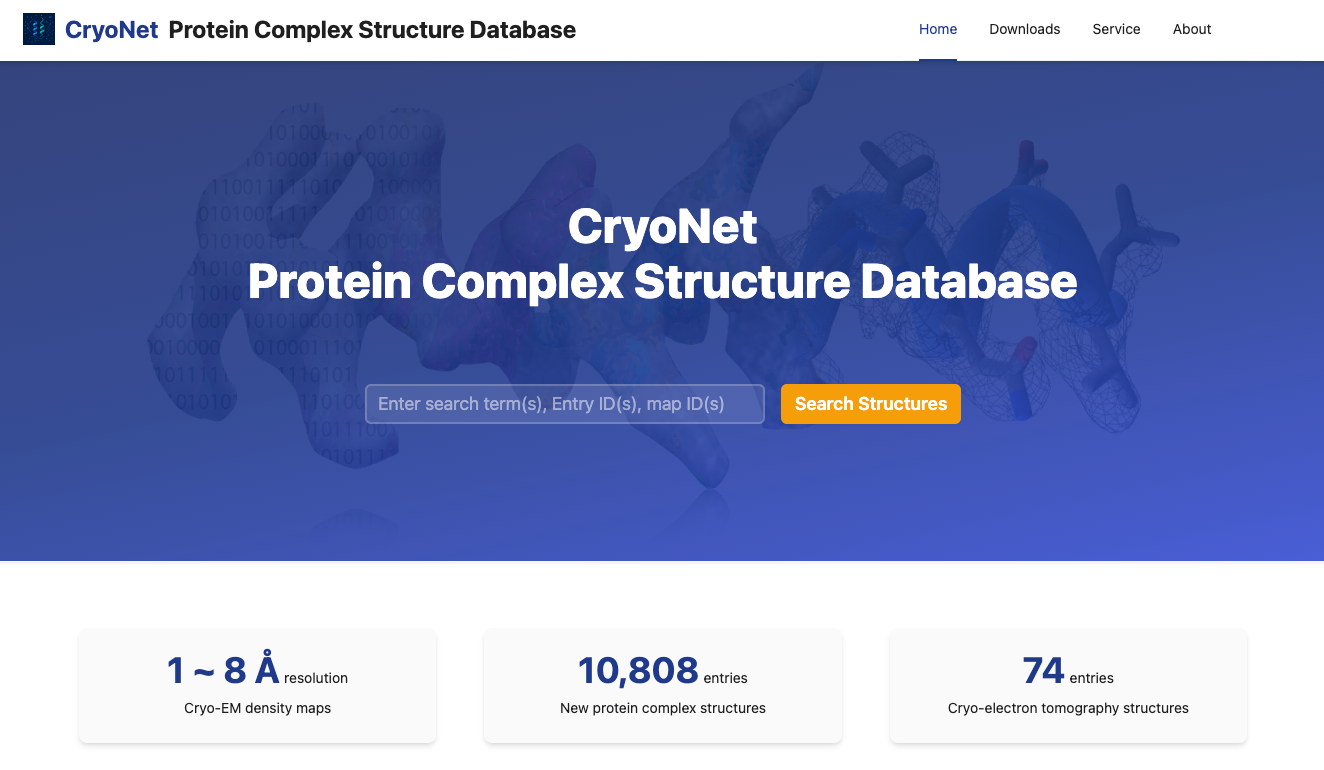
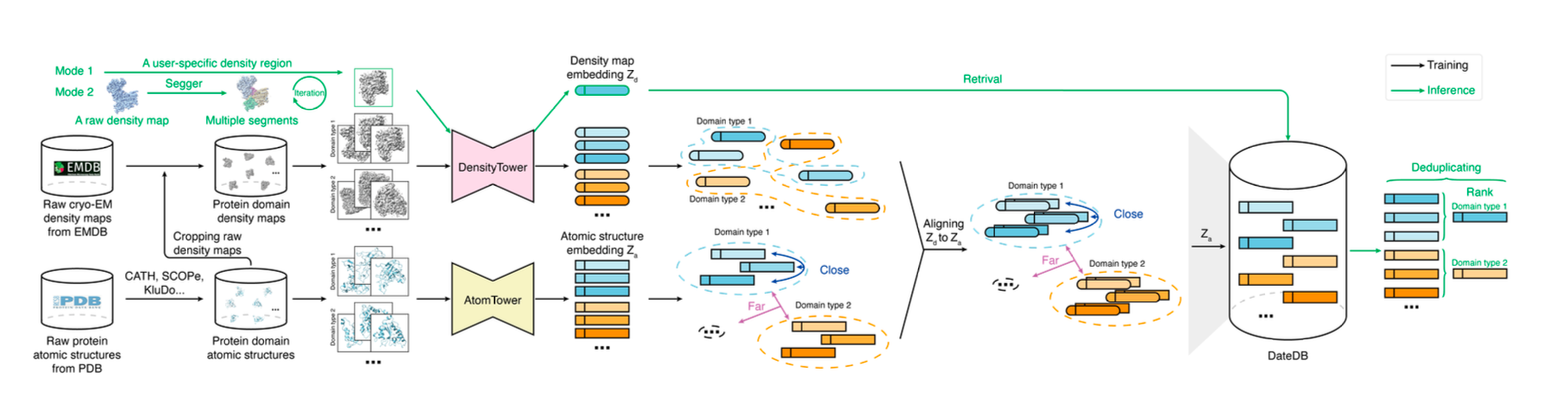
CryoNet.DB: AI-driven closed-loop biological structure database
Kui Xu*, Zhuoer Dong*, and Qiangfeng Cliff Zhang
CryoDomain: Sequence-free Protein Domain Identification from Low-resolution Cryo-EM Density Maps
Muzhi Dai, Zhuoer Dong, Weining Fu, Kui Xu#, and Qiangfeng Cliff Zhang#
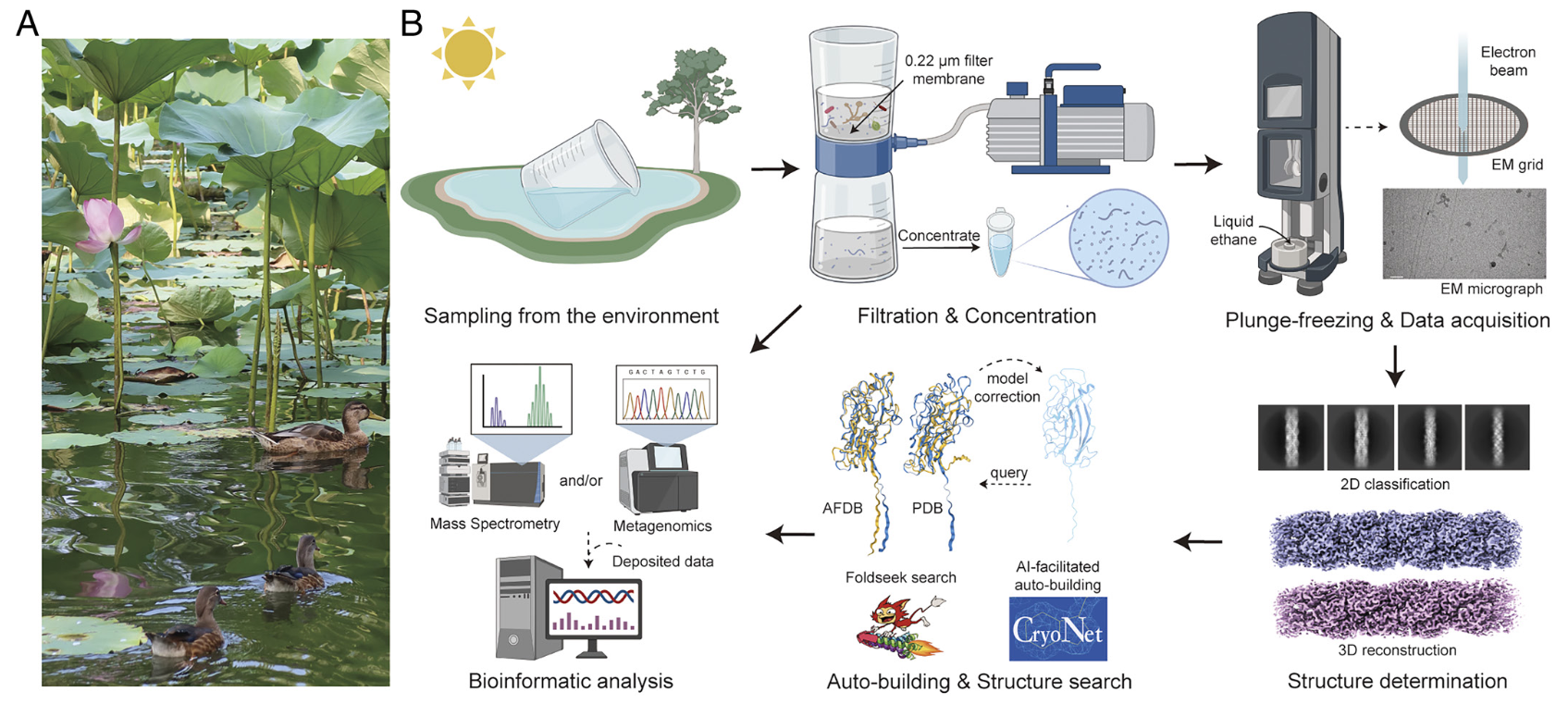
CryoSeek II: Cryo- EM analysis of glycofibrils from freshwater reveals well- structured glycans coating linear tetrapeptide repeats
Tongtong Wang*, Wenze Huang*, Kui Xu, Yitong Sun, Qiangfeng Cliff Zhang, Chuangye Yan#, Zhangqiang Li# , and Nieng Yan#
CryoSeek: A strategy for bioentity discovery using cryoelectron microscopy
Tongtong Wang*, Zhangqiang Li*,#, Kui Xu, Wenze Huang, Gaoxingyu Huang, Qiangfeng Cliff Zhang, and Nieng Yan#
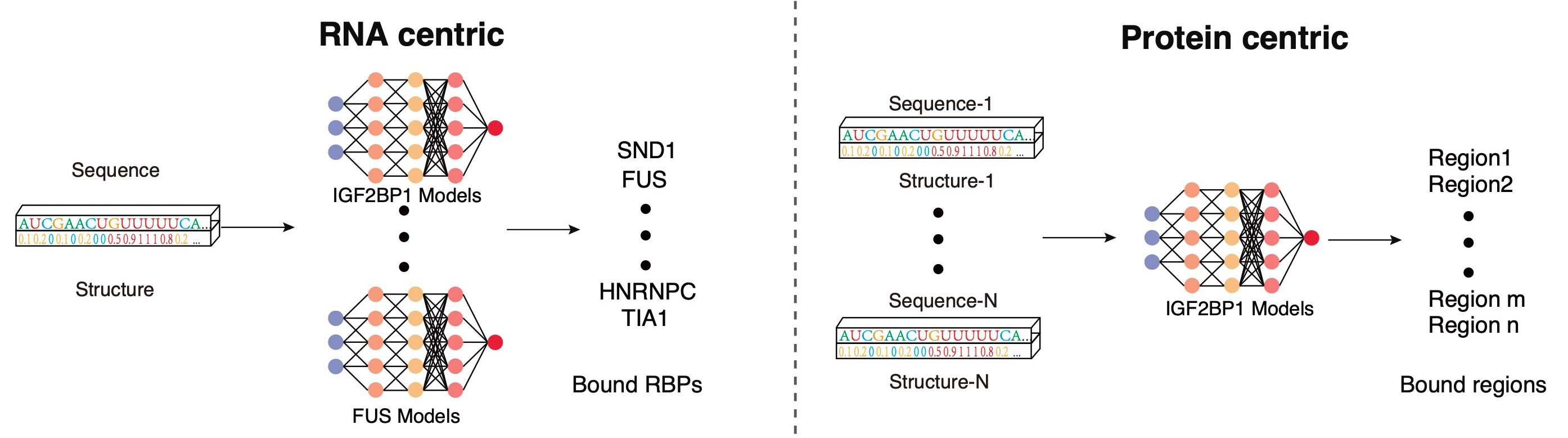
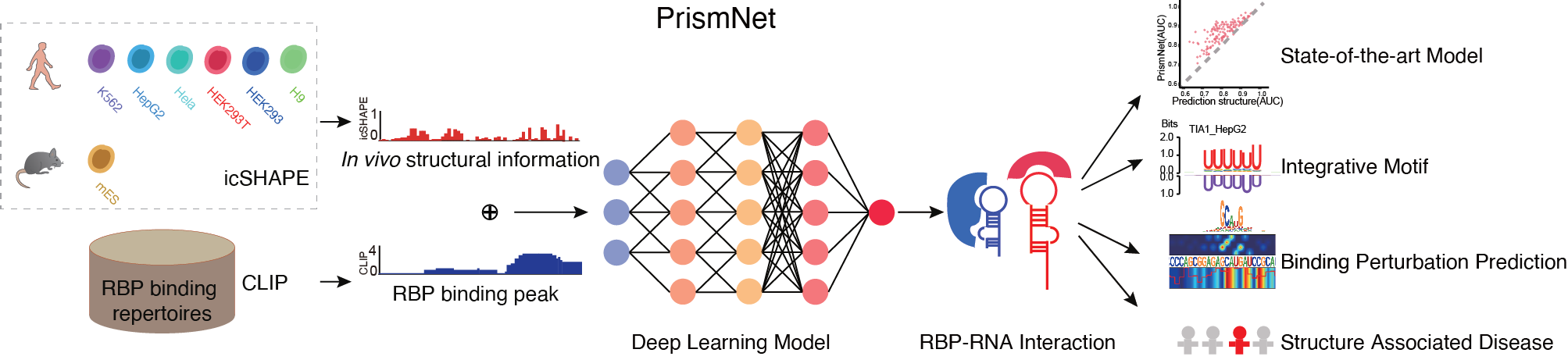
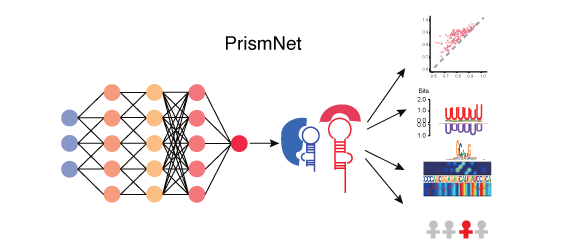
PrismNet: predicting protein–RNA interaction using in vivo RNA structural information
Yiran Xu*, Jianghui Zhu*, Wenze Huang*, Kui Xu, Rui Yang, Qiangfeng cliff Zhang#, Lei Sun#
Structural basis of membrane skeleton organization in red blood cells
Ningning Li#*, Siyi Chen*, Kui Xu, Meng-Ting He, Meng-Qiu Dong, Qiangfeng Cliff Zhang, and Ning Gao#
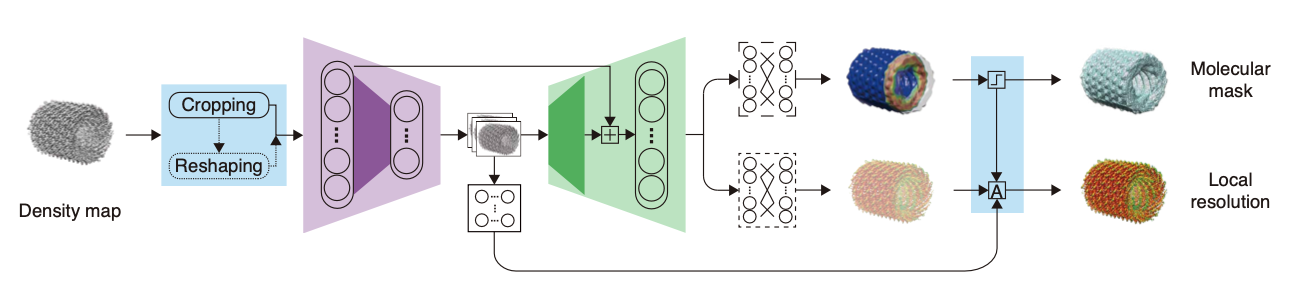
CryoRes: Local Resolution Estimation of Cryo-EM Density Maps by Deep Learning
Muzhi Dai, Zhuoer Dong, Kui Xu#, and Qiangfeng Cliff Zhang#
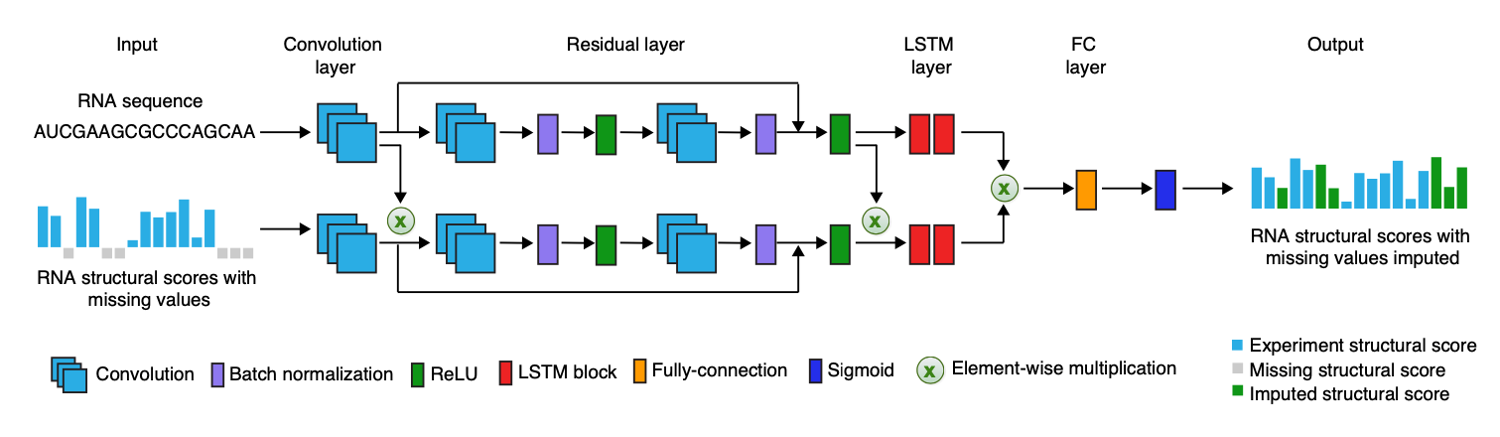
A deep learning method for recovering missing signals in transcriptome-wide RNA structure profiles from probing
experiments
Jing Gong*, Kui Xu*, Ziyuan Ma, Zhi John Lu and Qiangfeng Cliff Zhang
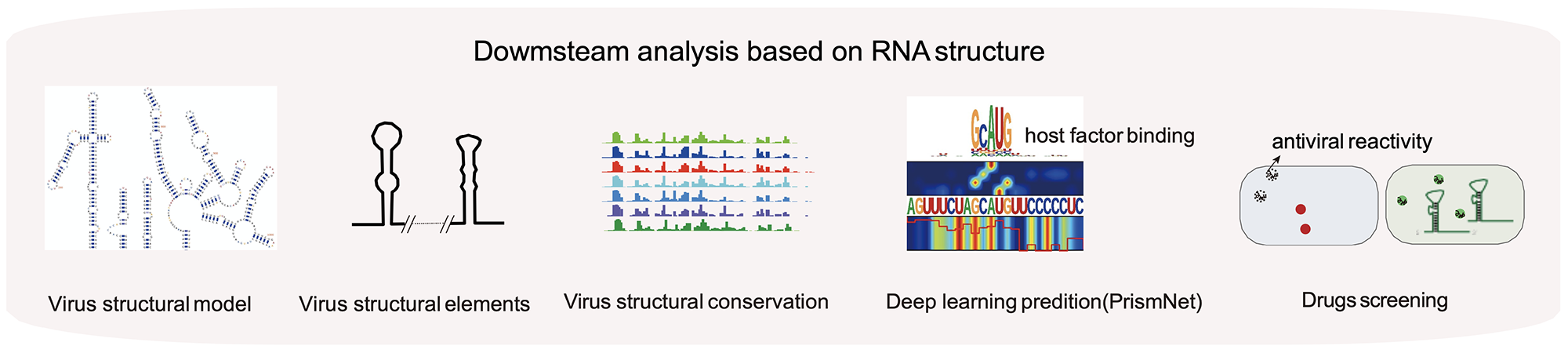
In vivo structural characterization of the whole SARS-CoV-2 RNA genome identifies host cell target proteins vulnerable to re-purposed drugs
Lei Sun*, Pan Li*, Xiaohui Ju*, Jian Rao*, Wenze Huang*, Shaojun Zhang, Tuanlin Xiong, Kui Xu, Xiaolin Zhou, Lili Ren, Qiang Ding#, Jianwei Wang# and Qiangfeng Cliff Zhang#
Predicting dynamic cellular protein-RNA interactions using deep learning and in vivo RNA structure
Lei Sun*, Kui Xu*, Wenze Huang*, Yucheng T. Yang*, Pan Li, Lei Tang, Tuanlin Xiong and Qiangfeng Cliff Zhang
Structure of the activated human minor spliceosome
Rui Bai*, Ruixue Wan*, Lin Wang, Kui Xu, Qiangfeng Cliff Zhang, Jianlin Lei, and Yigong Shi
RASP: an atlas of transcriptome-wide RNA secondary structure probing data
Pan Li*, Xiaolin Zhou*, Kui Xu, and Qiangfeng Cliff Zhang
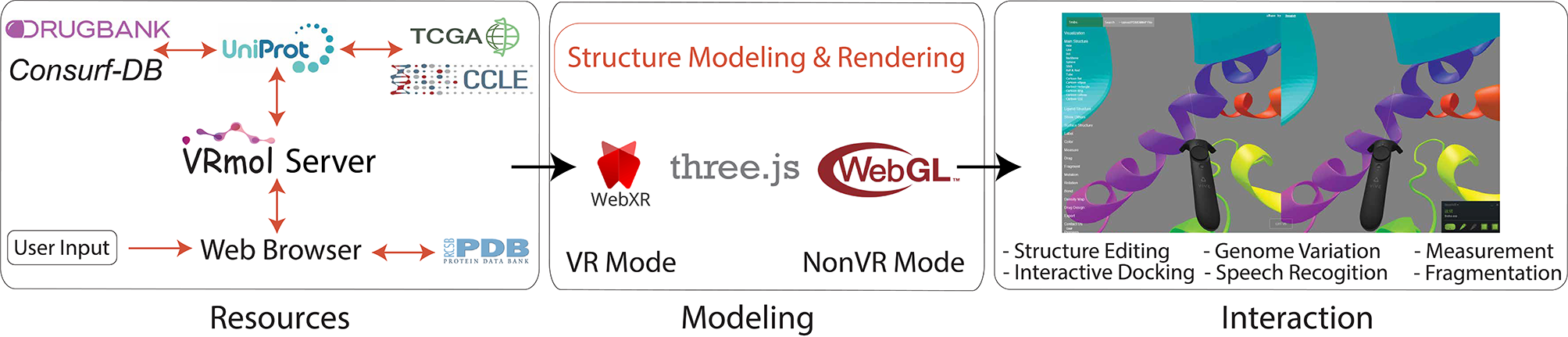
VRmol: an Integrative Web-Based Virtual Reality System to Explore Macromolecular Structure
Kui Xu*, Nan Liu*, Jingle Xu, Chunlong Guo, Lingyun Zhao, Hong-Wei Wang and Qiangfeng Cliff Zhang
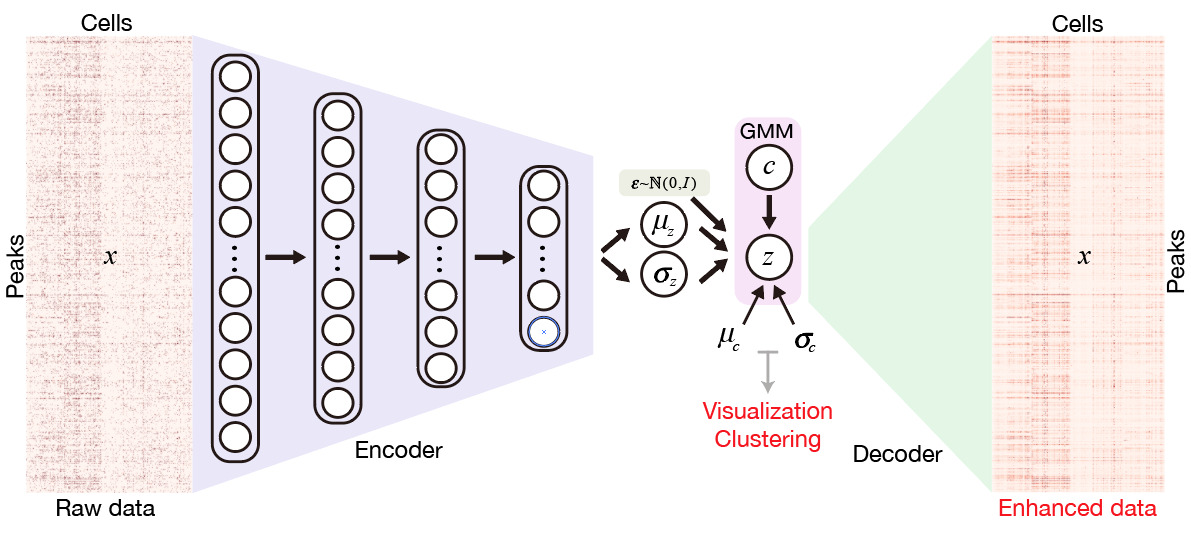
SCALE method for single-cell ATAC-seq analysis via latent feature extraction
Lei Xiong, Kui Xu, Kang Tian, Yanqiu Shao, Lei Tang, Ge Gao, Michael Zhang, Tao Jiang and Qiangfeng Cliff Zhang
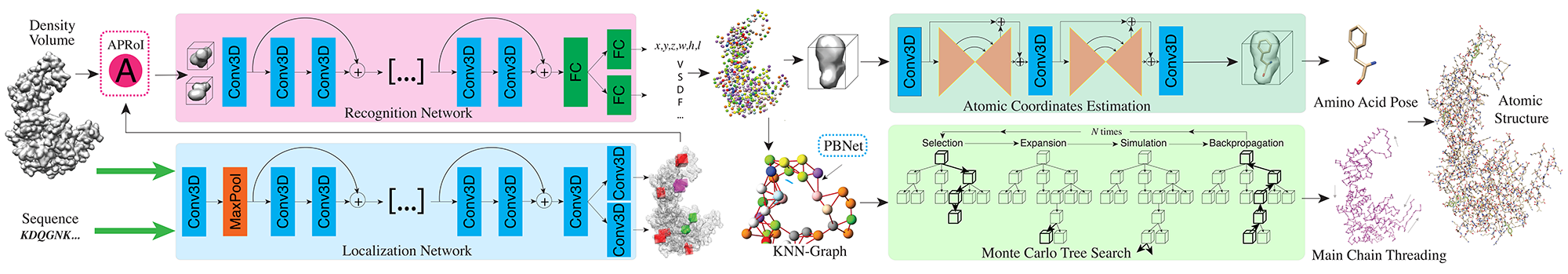
A2-Net: Molecular Structure Estimation from Cryo-EM Density Volumes
Kui Xu, Zhe Wang, Jianping Shi, Hongsheng Li and Qiangfeng Cliff Zhang
SegVoxelNet: Exploring Semantic Context and Depth-aware Features for 3D Vehicle Detection from Point Cloud
Hongwei Yi, Shaoshuai Shi, Mingyu Ding, Jiankai Sun, Kui Xu, Hui Zhou, Zhe Wang, Sheng Li and Guoping Wang
Bioactive functionalized monolayer graphene for high-resolution cryo-electron microscopy
Nan Liu, Jincan Zhang, Yanan Chen, Chuan Liu, Xing Zhang, Kui Xu, Jie Wen, Zhipu Luo, Shulin Chen, Peng Gao, Kaicheng Jia, Zhongfan Liu, Hailin Peng and Hong-Wei Wang
RISE: a database of RNA interactome from sequencing experiments
Jing Gong*, Di Shao*, Kui Xu, Zhipeng Lu, Zhi John Lu, Yucheng T. Yang and Qiangfeng Cliff Zhang
Reweighted sparse subspace clustering
Jun Xu, Kui Xu, Ke Chen and Jishou Ruan
(* co-First authors; # co-Corresponding authors)
Open Source
PrismNet, a deep learning tool that integrates experimental in vivo RNA structure data and RBP binding data for matched cells to accurately predict dynamic
RBP binding in various cellular conditions.
VRmol provides the visualization and analysis of macromolecule structures in an infinite virtual environment on the web. VRmol is natively built with
WebXR technology, providing functions in a fully immersive, inspiring virtual environment.
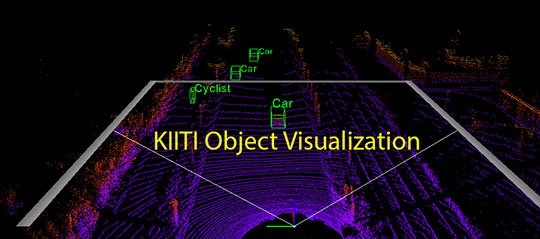
Kitti Object Visualization
Data Transformation and Visualization for
Kitti Object Detection Dataset, including mapping 3D boxes on LiDar point cloud in
volumetric mode, mapping 2D and 3D boxes on Camera image, mapping 2D boxes on LiDar Birdview and mapping LiDar data on Camera image.
Implementing a 3D
DenseNet for 3D CAD Object Classification on
ModelNet dataset using highly
efficient
Torch. VoxNet, C3D, and 3D-VGG models are also provided. Multi-GPU training is well supported. Check
ModelNet Benchmark Leaderboard to see related researches.
A collaborative list of 3D Deep Learning works via 3D Representation, 3D Classification, 3D Classification, 3D Object Detection, 3D Reconstruction & Generation, 3D Human Pose Estimation and their related datasets.
A collaborative list of awesome CryoEM (Electron Cryo-Microscopy) resources, including Methods and Softwares related Particle Picking, 3D Classification, Denoising, Motion Correction, 3D Reconstruction, Model Building and some other related research.
V. Falconieri, S. Subramaniam, NCI-NIH